23. Export Your Data¶
23.1. Raw Data Export¶
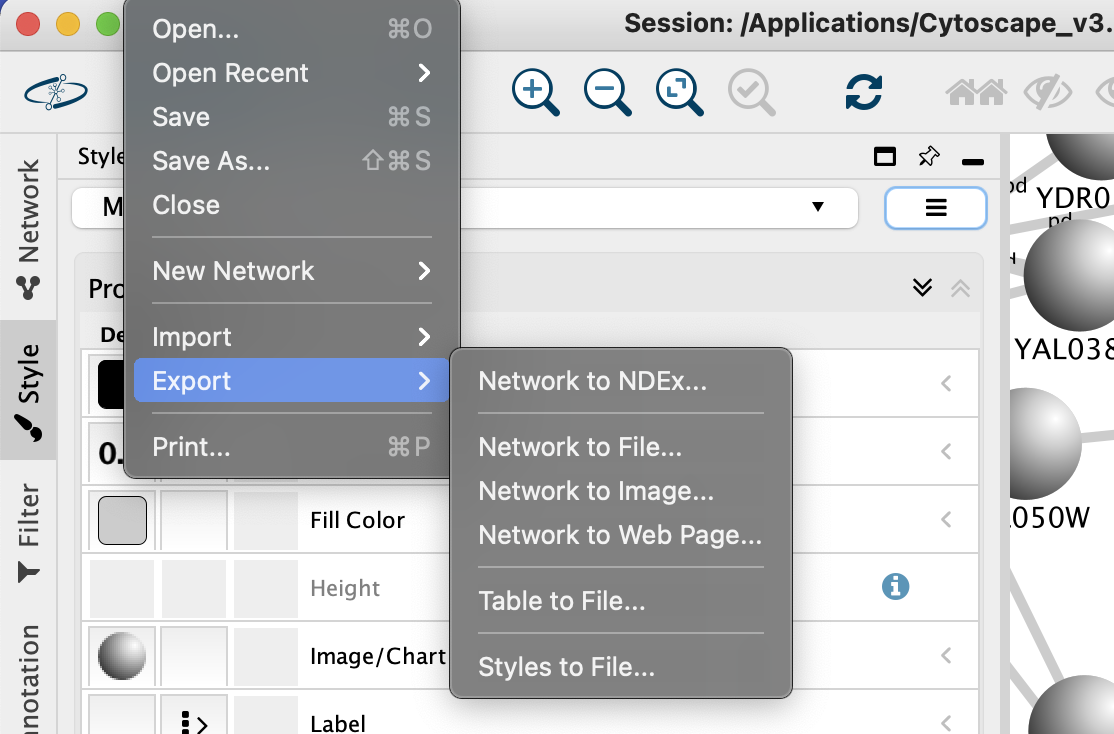
To use data from Cytoscape networks in an external application, the data has to be exported. As a Cytoscape network consists of a number of different types of data, there are options in Cytoscape for each exportable data type. These options (Network, Table, Styles) can be found on the File → Export submenu (shown above).
Network Data Exchange (NDEx) Public Database¶
The Network to NDEx option is used to store a network on the public NDEx server. You can use the NDEx public server to store your networks on the cloud, for sharing networks with collaborators, or to archive networks for publication. To store a network, you must first create a free NDEx account, which you can easily do by visiting the NDEx Public Server website.
The Network to NDEx option saves only the currently selected network to your NDEx account. Such networks are suitable for processing by other applications and web services, as they contain only the nodes and edges in the selected network – other views and the nodes and edges unique to them are not written. If you later re-import this network (using the Cytoscape Search bar), only the single network you saved will be loaded.
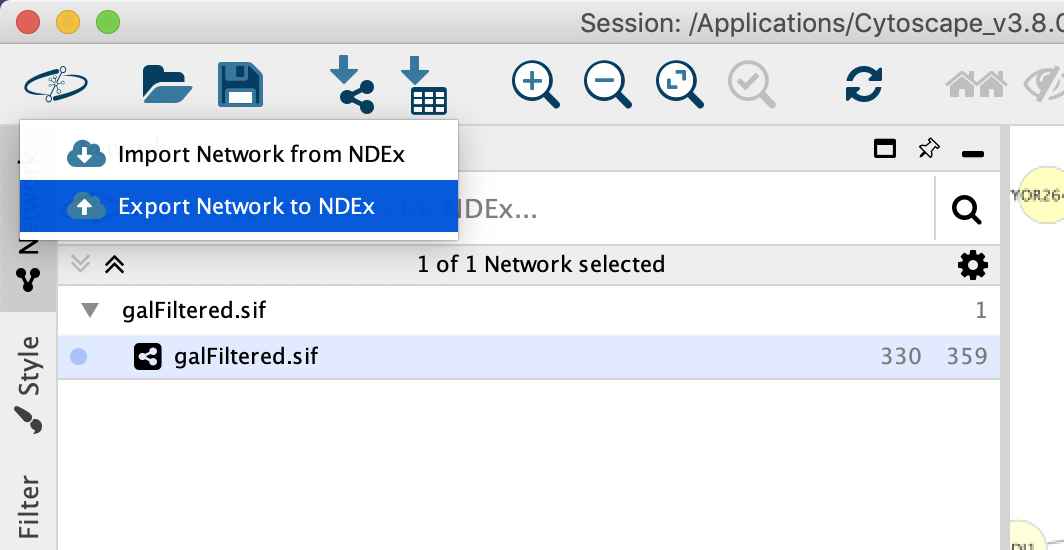
When you select the option, the CyNDEx-2 Browser dialog will appear and enable you to provide various metadata to annotate the network and make it findable in subsequent NDEx searches. You can set the name of your network by editing the Network Name attribute. The Update Existing Network option enables you to overwrite the existing copy of the network (if you have permission to do so). We recommend to provide as much metadata as possible, and always provide at least Version and Description attributes.
To export your network, you must first associate your NDEx account credentials with the CyNDEx-2 Browser by filling out the profile form (red oval in screenshot below).
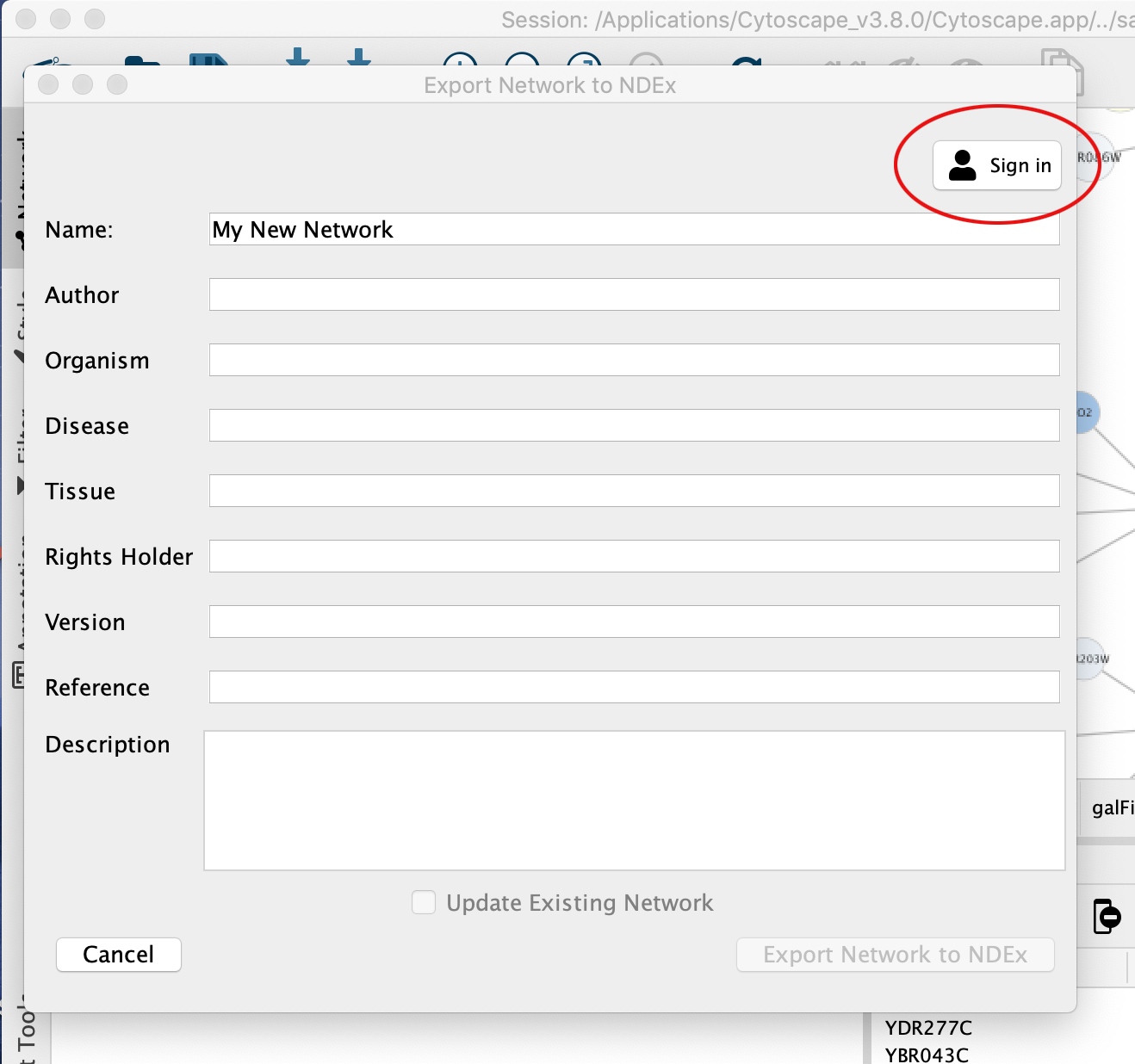
Once the profile is created, you are allowed to export your network to NDEx.
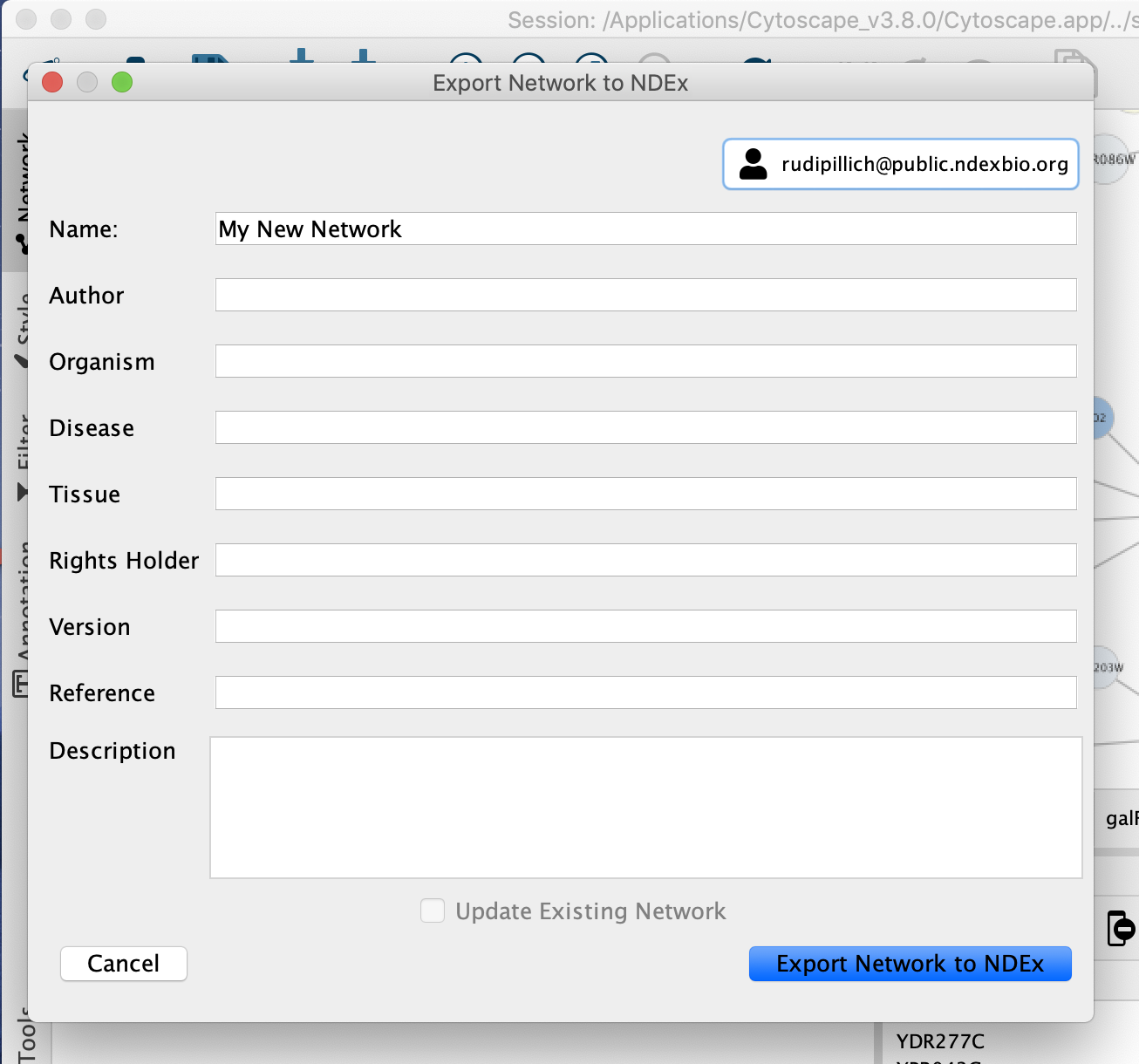
Export commands have also been added to the network and collection context menus in the Cytoscape Network panel, as another method of accessing the network save dialog.
Finally, the NDEx button in the toolbar provides an additional convenient way to both Import and Export to NDEx.
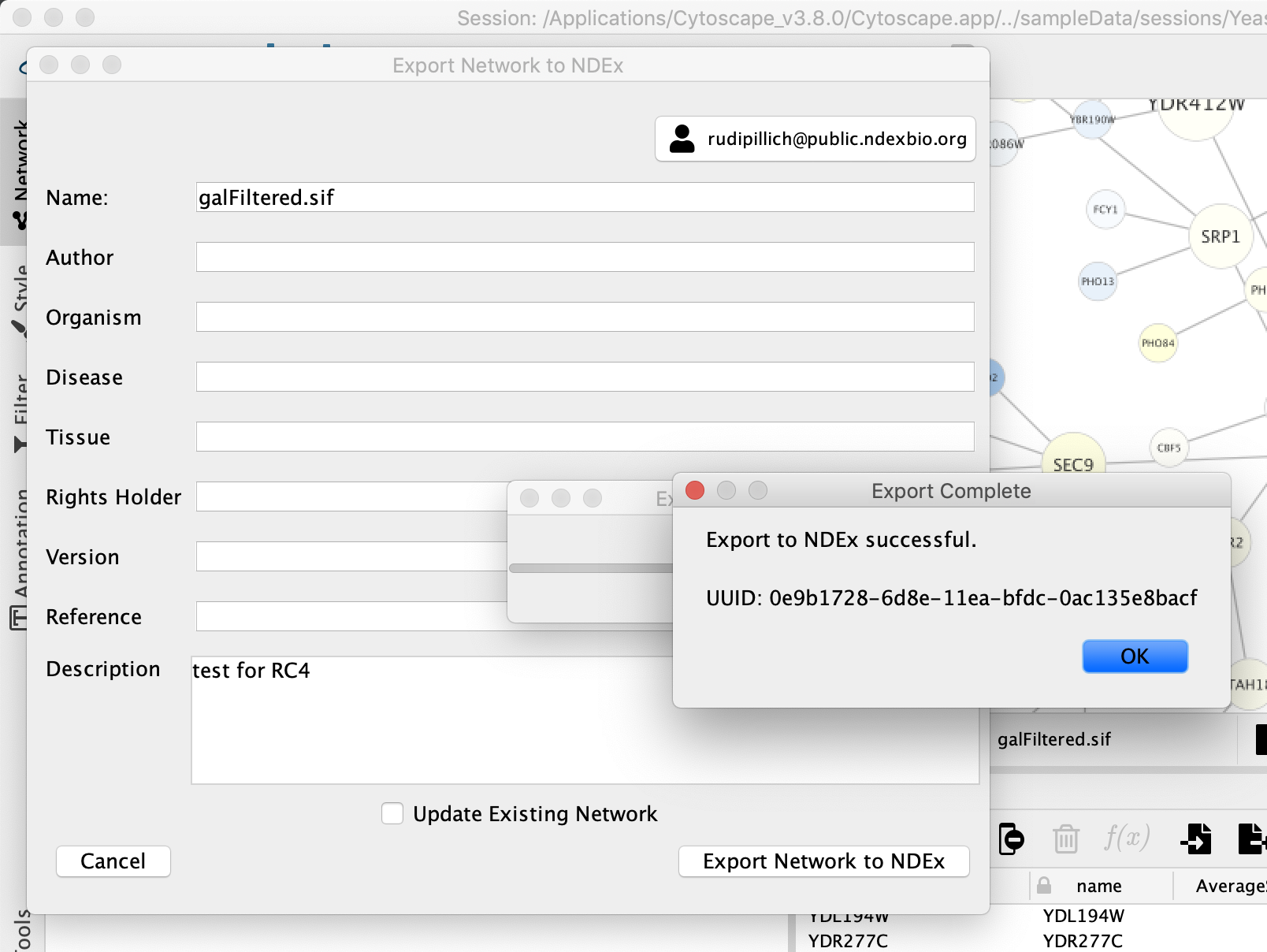
When the export operation is complete, a confirmation dialog will be displayed as shown below. If you want to share your network with external collaborators or for more sharing options, please refer to the guide for Uploading and Sharing Networks in NDEx.
Further information on NDEx is available at the NDEx Informational/Doc website.
Network¶
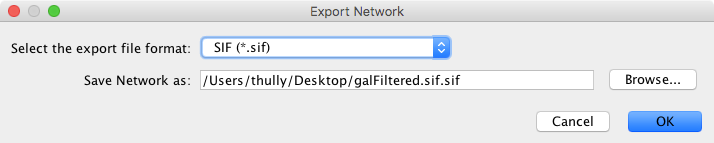
The Network option is used to export network data. Network data includes the nodes and edges in the network, and also will include data attributes and visual information if the chosen format supports it (like XGMML). Network data can be exported in a number of formats, including:
SIF (Simple Interaction Format)
XGMML, with or without style data
GraphML
PSI-MI Level 1 and 2.5
Cytoscape.js JSON (can be used with Cytoscape.js, a tool described in future sections)
CX JSON (for Cytoscape Cyberinfrastructure network exchange)
CX2 JSON
The default format is SIF, which is a simple tab-delimited network format that provides node names and edge interactions only. You can choose the format using the dialog drop-down box. A file name is automatically suggested based on the network name and selected format, with the file path defaulting to the current working directory. You can change the suggested file name/path using the text box or Browse button. Press OK to export with the selected options.
Table¶
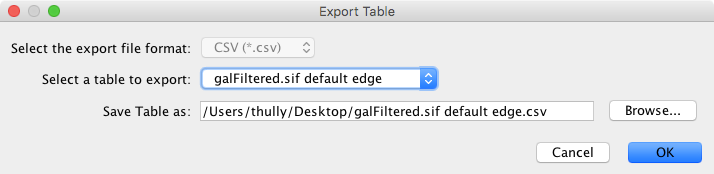
The Table to File… export option is used to export any of the data tables that are available in the current Cytoscape session. This includes the node, edge, and network tables that can be seen in the Table Panel, as well as any unassigned tables. The only currently supported file format currently for export is CSV (comma-separated value).
The export dialog for Table has only two selectable options - the table to export and the file name (file type is always CSV). A file name is suggested based on the name of the selected table with the path defaulting to the current working directory. As in Network, the suggested file name can be changed using the Browse button or the text field. Press OK to export the selected table with the given file name.
A shortcut button for Export Table to File… is also available in the Table Panel
 .
.
Styles¶
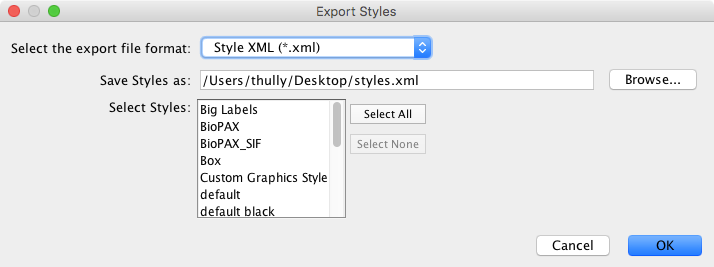
The Styles option is used to export one or more styles available in the current Cytoscape session. Supported formats include Style XML (which can be imported by 3.x versions of Cytoscape) or Cytoscape.js JSON (which can be used with Cytoscape.js, described in future sections).
Style export has three options - the export format (as described above), the style(s) to export, and the file name (a suggestion will be made based on the current directory and file type). As in Network → Table, the suggested file name can be changed using the Browse button or the text field. The list of styles is a multiple-selection list - one or more can be selected using Ctrl/Cmd-click (to select multiple discontiguous elements) or Shift-click (to select a contiguous element range). Press OK to export all selected styles with the given file path.
23.2. Exporting for Publication¶
When you finish your data analysis and visualization, you can publish your data to share the results. Cytoscape has several options to do this, with most options suitable for Cytoscape users and other options suitable for programmers wanting to create unusual or complex network viewers. These are further explained below.
An NDEx network
A session file
A static image
An interactive web application
Full web application
Simple network view (for web application developers)
As an NDEx Network¶
The easiest way to share your results with others is to save your network in the NDEx Public Database by using the Network to NDEx or Collection to NDEx options. Then, visit your NDEx account and enable the Sharable URL (similar to a Google Doc shared link) so that your private network can be accessed by whoever you send the link to.
NDEx also has additional options that enable you to:
request a DOI suitable for use in publications, grant applications or progress reports
create working groups and specific sharing within NDEx.
run different types of queries on your networks
perform enrichment analysis on a variety of datasets, including curated pathways, large interactomes, drug-target association networks and many more.
For more information on NDEx publishing and sharing features, see the Publishing in NDEx and Sharing Networks in NDEx manuals.
As a Session File¶
Another easy way to share your results with others is simply saving everything as a session file (which is a zipped session archive). You can save your current session by clicking the Save Session icon. You can save to a thumb drive, a shared file system, or even a cloud drive directory such as Dropbox. If you save to a shared drive, beware not to have two people work on the same session file with Cytoscape at the same time, as unpredictable results may occur.
As a Static Image¶
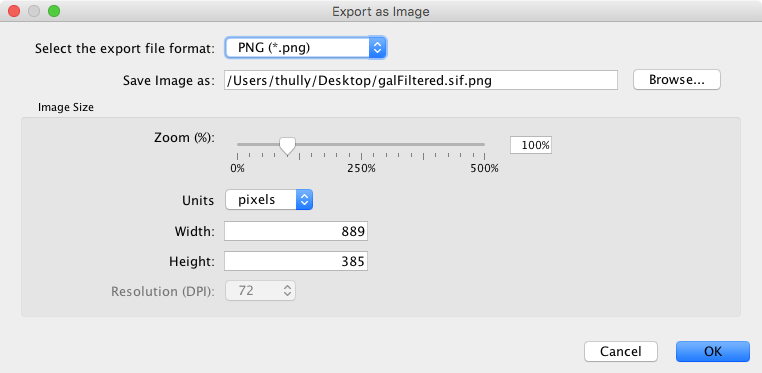
Cytoscape can generate publication-quality images from network views. By selecting File → Export as Image…, you can export the current network view into the following formats:
JPG
PNG
PS (Post Script)
SVG
PDF
The image export dialog has a variable number of options based on the file format selected, with file type and file name always being selectable. PNG and JPEG have options for scaling, while other formats only have a single option, Export Text as Font. The file name is suggested based on the name of the selected table with the path defaulting to the current working directory. As in raw data export, the suggested file name can be changed using the Browse button or the text field. Press OK to export an image with the given file name and parameters.
We recommend using PDF for publications because it is a standard vector graphics format, and it is easy to edit in other applications such as Adobe Illustrator.
Known Issues¶
For PDF export, there is an option to Export text as font. This option does not work for two-byte characters such as Chinese or Japanese. To avoid corrupted texts in the exported images, please uncheck this option when you publish networks including those non-English characters.
As an Interactive Web Application¶
The Web is an excellent platform for data sharing and collaboration, and Cytoscape (as of version 3.2.0) provides a number of ways to publish your network on the web, with each choice representing tradeoffs between ease, simplicity, and customization options. All solutions leverage the cytoscape.js drawing library, and so enable not only viewing but also Cytoscape-style interactive browsing of networks and attributes.
The simplest choice is CyNetShare, where you save your network (and optionally a style) on a public file system, then interactively view the network in a browser. Like Google Maps, you can generate and publish a URL that allows collaborators to also view your network.
Alternatively, Cytoscape can generate an entire web site showing a single page containing the viewer with your network pre-loaded. You can load this directly onto your own web server to become part of your web site.
Finally, Cytoscape can generate the skeleton of a web site for further customization by JavaScript programmers.
These features are available as Export menu items under the File menu, and are described in sections below.
For example, here is a network in Cytoscape:
Here is the same network as an interactive web visualization:
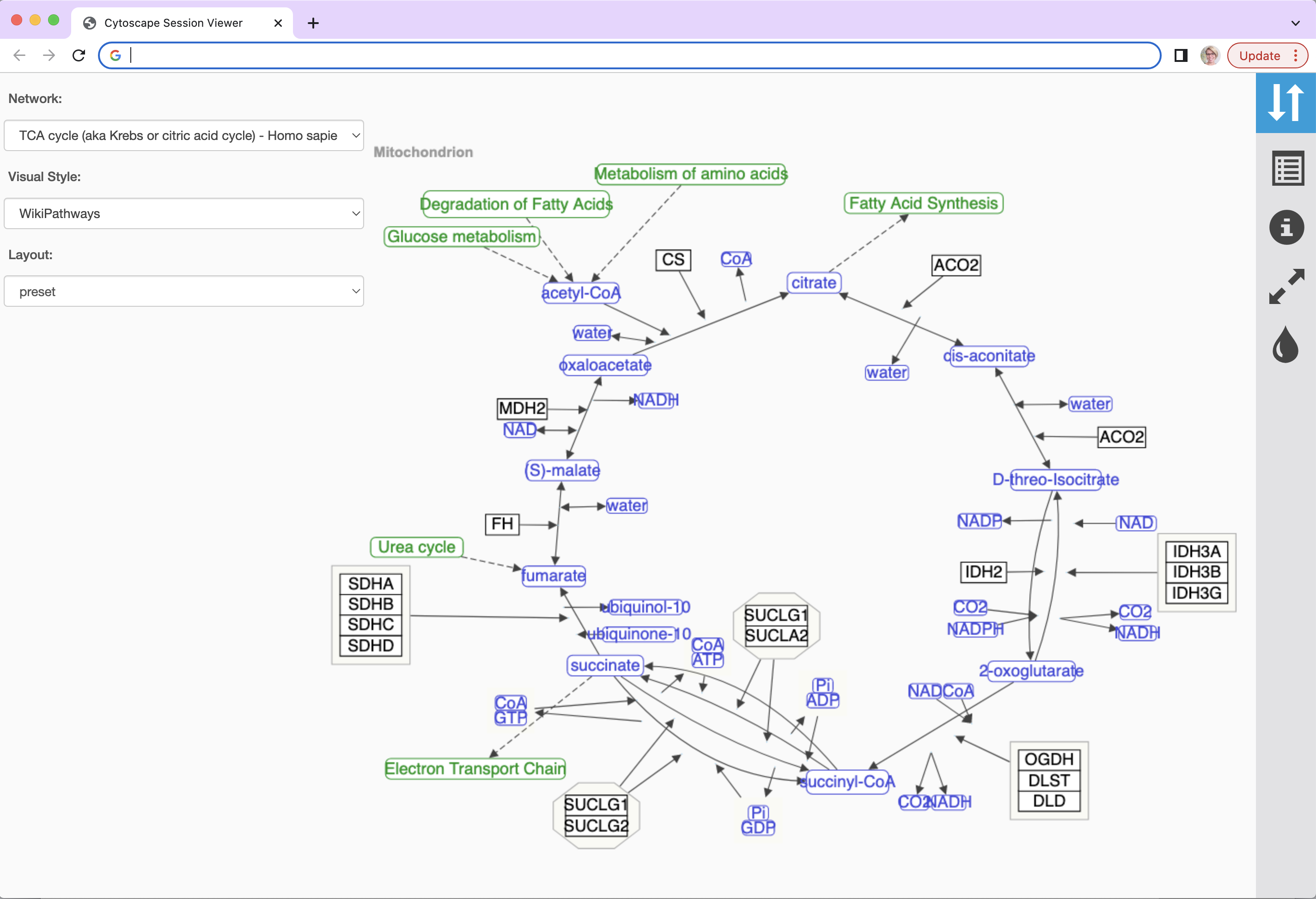
Note that web browsers can render small networks (e.g., 1000 nodes) quickly and effectively, but attempting to render large ones (e.g., 5000 nodes) will take a very long time.
A word about exporting styles to interactive web applications: Our web applications are based on the cytoscape.js display library, which renders a subset of Cytoscape styles. For more information, see the Export Styles to Cytoscape.js section below.
Generating a Full Web Application¶
The full page export option is designed for users who want to publish their network as a complete single-page application. Cytoscape creates a zip archive containing a complete JavaScript-based web application that works as a basic viewer (like CyNetShare) for Cytoscape-generated network visualizations. You can unzip the archive onto a web server (or your PC) and view the network via a web browser on PCs and tablets.
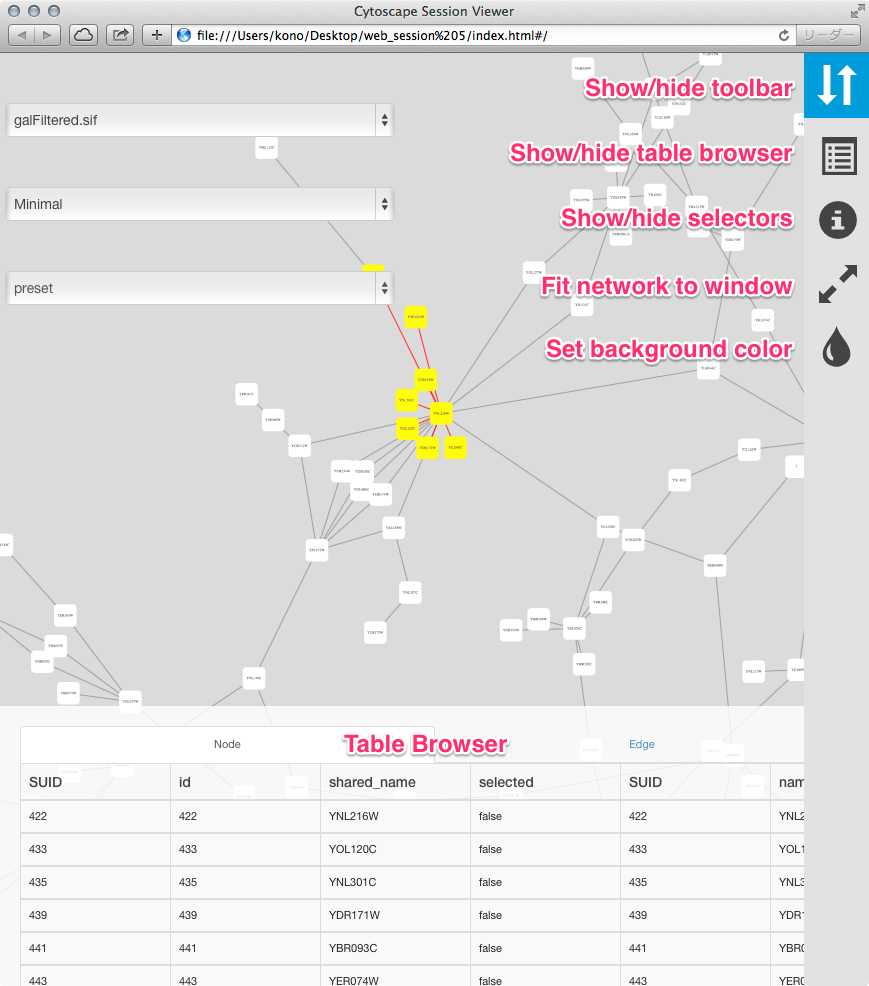
To generate an entire web page for all networks in the session as a zip archive, select File → Export → Network to Web Page…. The following dialog will appear:
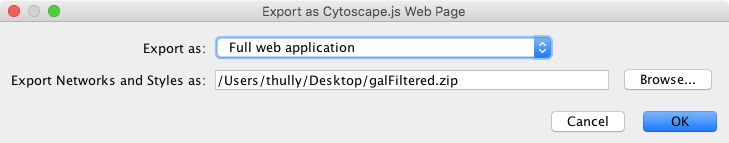
This simple dialog has only two options - the file name and the type of web export (full, simple network view, or JSON). For full web application, we use the default. The file name is suggested based on the name of the selected table with the path defaulting to the current working directory. As in raw data export, the suggested file name can be changed using the Browse button or the text field. Press OK to export a web archive with the given file name.
To view the web page, unzip the archive into a folder on your PC or web server. The folder will contain an index.html file, the network data, and other files. You can open the index.html file in your browser (usually from your browser’s File → Open menu item.)
Here is an example exported file from Cytoscape:
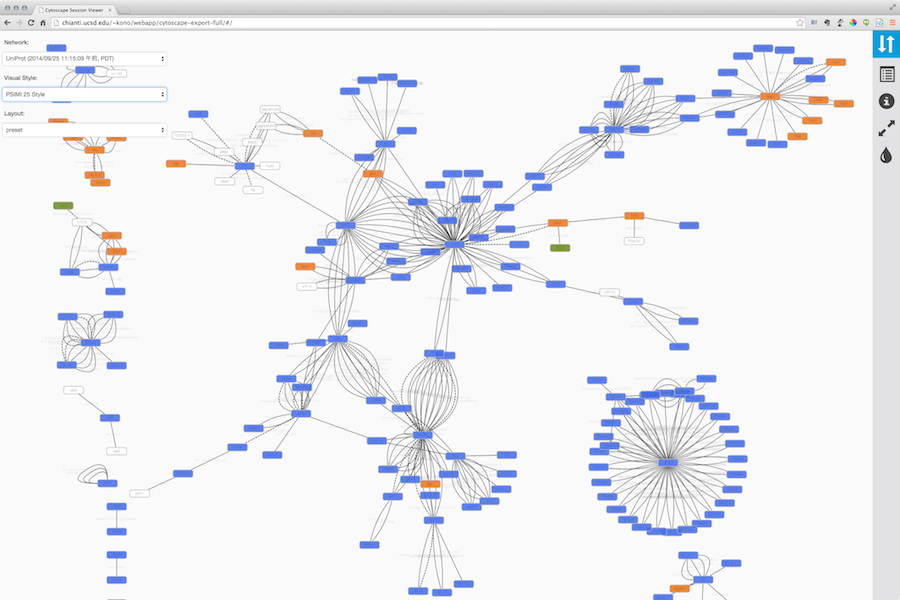
Note that because Cytoscape uses the latest HTML5-based web technologies, it cannot support older or non-conformant web browsers such as Internet Explorer. We strongly recommend using the latest version of modern web browsers such as Google Chrome, Mozilla Firefox, or Apple Safari.
Generating a Simple Network View (For Web Application Developers)¶
The Simple Network View export option is designed for users who want to publish their data as a complete single-page application, but are interested in customizing the web viewer application themselves. Cytoscape creates a zip archive containing a partial JavaScript-based web application based on the cytoscape.js library and including simple boilerplate code and the current network view. The user can create a custom viewer by customizing this code.
To generate a web page for a single network view as a zip archive, select File → Export as Web Page … as before, but this time choose the Simple viewer for current network only format as below:
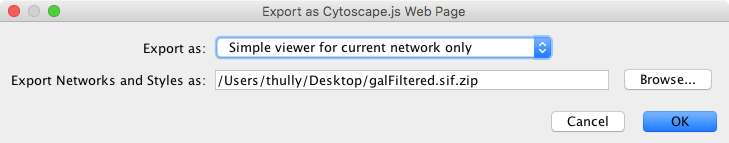
Everything else works as before, though this time the viewer will only include the current network. Press OK to proceed with the export.
For instructions on testing the customized web application, see Generating a Full Web Application above.
Extending the webpage with Cytoscape.JS functionality¶
As of version 3.6.2 of the JSON Support core app in Cytoscape, the user may add custom Cytoscape.JS events to the exported webpage. In the web_session/scripts directory, a custom.js file has been added with a designated spot for custom JS code.
For example, to add linkouts to nodes in the webpage (i.e. opening a URL specified by the href node attribute when the user clicks the node), the custom.js file should look like:
// Notice that the `cy` object may only be referenced
// after the document has finished loading.
$( document ).ready(function(){
cy.on('tap', 'node', function(){
try { // your browser may block popups
window.open( this.data('href') );
} catch(e){ // fall back on url change
window.location.href = this.data('href');
}
});
});
This change has to be made each time you export a web page. If you want to change the default export template, refer to the next section.
Customize Export Template (For Web Application Developers)¶
The code generated by Cytoscape for the Full Web Application and the Simple Network View web applications is minimalistic. While you can directly modify this code yourself to create your own page design or add new features, the modifications will apply to a single exported network. If you are a web application developer, you can change the application code generated for all exports by editing HTML5 template code resource files in your ~/CytoscapeConfiguration/web directory:
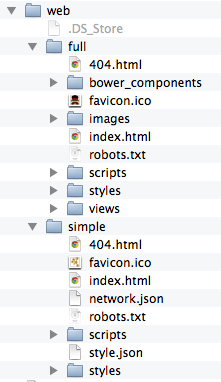
In this folder, you can find full and simple sub directories corresponding to Full Web Application and the Simple Network View described above.
Requirements¶
To build these project, you need the following tools installed:
Node.js
gulp
grunt
Full Export Template¶
This is an AngularJS based web application built with grunt (at least for now – we have plans to migrate to gulp). Source code and more documentations are available here:
To build the project into dist directory, type:
grunt
Simple Export Template¶
This template is generated by a simple gulp project. The source code is available here:
To build the project into dist directory, type:
gulp
Use Your Custom Templates for Export¶
Once you have your own builds, you can deploy your templates by replacing the contents of full and simple with your own builds.